 |
NAMD has a number of options that control the way that non-bonded interactions are calculated. These options are interrelated and can be quite confusing, so this section attempts to explain the behavior of the non-bonded interactions and how to use these parameters.
 |
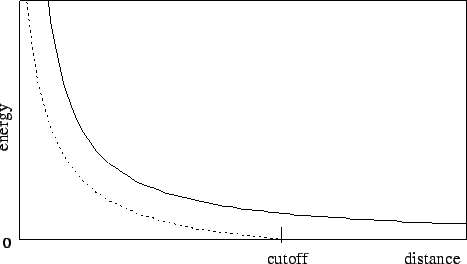
The switching function used is based on the X-PLOR switching function. The parameter switchdist specifies the distance at which the switching function should start taking effect to bring the van der Waals potential to 0 smoothly at the cutoff distance. Thus, the value of switchdist must always be less than that of cutoff.
First let us consider the latter case, where electrostatics are truncated at the cutoff distance. Using this scheme, all electrostatic interactions beyond a specified distance are ignored, or assumed to be zero. If switching is set to on, rather than having a discontinuity in the potential at the cutoff distance, a shifting function is applied to the electrostatic potential as shown in Figure 2. As this figure shows, the shifting function shifts the entire potential curve so that the curve intersects the x-axis at the cutoff distance. This shifting function is based on the shifting function used by X-PLOR.
 |
Next, consider the case where full electrostatics are calculated. In this case, the electrostatic interactions are not truncated at any distance. In this scheme, the cutoff parameter has a slightly different meaning for the electrostatic interactions -- it represents the local interaction distance, or distance within which electrostatic pairs will be directly calculated every timestep. Outside of this distance, interactions will be calculated only periodically. These forces will be applied using a multiple timestep integration scheme as described in Section 5.2.
 |
Let us consider a concrete example to better understand this. Assume that the pairlist is built once every ten timesteps and that the cutoff distance is 8.0 Å. Consider a pair of atoms A and B that are 8.1 Å apart when the pairlist is built. If the pair list includes only those atoms within the cutoff distance, this pair would not be included in the list. Now assume that after five timesteps, atoms A and B have moved to only 7.9 Å apart. A and B are now within the cutoff distance of each other, and should have their non-bonded interactions calculated. However, because the non-bonded interactions are based solely on the pair list and the pair list will not be rebuilt for another five timesteps, this pair will be ignored for five timesteps causing energy not to be conserved within the system.
To avoid this problem, the parameter pairlistdist allowed the user
to specify a distance greater than the cutoff distance for pairs
to be included in the pair list, as shown in Figure 4.
Pairs that are included in the pair list but are outside the cutoff distance
are simply ignored. So in the above example, if the pairlistdist
were set to ![]() Å, then
the atom pair A and B would be included in the pair list, even though
the pair would initially be ignored because they are further apart than
the cutoff distance. As the pair moved closer and entered the cutoff
distance, because the pair was already in the pair list, the non-bonded
interactions would immediately be calculated and energy conservation
would be preserved. The value of pairlistdist should be chosen
such that no atom pair moves more than
Å, then
the atom pair A and B would be included in the pair list, even though
the pair would initially be ignored because they are further apart than
the cutoff distance. As the pair moved closer and entered the cutoff
distance, because the pair was already in the pair list, the non-bonded
interactions would immediately be calculated and energy conservation
would be preserved. The value of pairlistdist should be chosen
such that no atom pair moves more than
![]() in one cycle. This will insure energy conservation.
in one cycle. This will insure energy conservation.
 |
NAMD 2.X eliminated the explicit use of pairlists in order to reduce memory usage in light of equally efficient distance-testing algorithms. Specifically, it was realized that building a pairlist on top of the existing spatial decomposition was only marginally more efficient than actually testing atom distances at every timestep given efficient methods for dealing with nonbonded exclusions. The pairlistdist parameter now serves the same function, but is instead used to determine the minimum patch size. Unless the splitPatch parameter is explicitly set to position, hydrogen atoms will be placed on the same patch as the ``mother atom'' to which they are bonded. These hydrogen groups are then distance tested against each other using only a cutoff increased by the the value of the hgroupCutoff parameter. The size of the patches is also increased by this amount. NAMD functions correctly even if a hydrogen atom and its mother atom are separated by more than half of hgroupCutoff by breaking that group into its individual atoms for distance testing. Warning messages are printed if an atom moves outside of a safe zone surrounding the patch to which it is assigned, indicating that pairlistdist should be increased in order for forces to be calculated correctly and energy to be conserved.