


Next: Analysis Scripts
Up: Timeline Tutorial
Previous: Interface and controls
Contents
Subsections
Trajectory analysis: titin domain extension
To use Timeline to identify events in a trajectory, we must choose
which parameters to examine, perform the required analyses, and explore the
resulting 2D data sets. We use as an example the titin I91 extension trajectory
introduced above, and start with examining secondary structure, to get an
overall sense of the architectural changes that take place during forced
extension. Then we will look at geometric fluctuations during the trajectory,
which will both illustrate how to use some of the Timeline tools and
provide some hints as to when and where important events are taking place in
the I91 extension. Then we will look at the hydrogen bond breaking between the
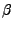 -strands of the domain's
-strands of the domain's 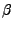 -sheets, to get more insight into how
the protein architecture handles applied force.
-sheets, to get more insight into how
the protein architecture handles applied force.
We examine how the secondary structure of the domain changes as the force
extension takes place. We can see visually by animating the 3D trajectory that
the domain begins as a 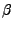 -sandwich then unravels as the force extension
takes place, but there is more to the story; we can make a 2D data plot to
examine the fate of each
-sandwich then unravels as the force extension
takes place, but there is more to the story; we can make a 2D data plot to
examine the fate of each 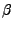 -strand.
-strand.
- Start a new VMD session. Launch VMD by:
- Unix/Mac OS X Users:
typing vmd in a Terminal window.
- Windows Users:
using the Start menu.
- Load in the PSF file for titin I91 domain. In the VMD Main window, select File
 New Molecule, then in the Molecule File Browser window, with Load files for: New Molecule selected, navigate to the timeline-tutorial-files directory and select titin.psf.
New Molecule, then in the Molecule File Browser window, with Load files for: New Molecule selected, navigate to the timeline-tutorial-files directory and select titin.psf.
- Load in the titin extension trajectory. In the Molecule File Browser window, with Load files for: 0: titin.psf selected, open titin.dcd, which is also in the timeline-tutorial-files directory. The trajectory shows a steered molecular dynamics extension of the I91 domain of the giant muscle protein titin, solvated in a water droplet with a non-periodic boundary.
- Adjust the VMD Main time slider to frame 0. In the VMD Main window, select Graphics
 Representations... to display the Graphical Representations window. In the Graphical Representations window, edit the default all selection to not water. Set the Coloring Method to Secondary Structure and Drawing Method to New Cartoon.
Representations... to display the Graphical Representations window. In the Graphical Representations window, edit the default all selection to not water. Set the Coloring Method to Secondary Structure and Drawing Method to New Cartoon.
- Open the Timeline plugin window. In the VMD Main window, select Extension
 Analysis
Analysis  Timeline.
Timeline.
- Display secondary structure for all frames, all residues in the trajectory. In the VMD Timeline window, select Calculate
 Calc. Sec. Structure
Calc. Sec. Structure
- Scrub the cursor and explore the 2D data plot. With left mouse depressed move the 2D cursor around the 2D data plot. Note how the 3D highlight (a red Licorice representation) moves around the molecule, highlighting whatever is currently under the 2D cursor.
Figure:
Secondary structure for all residues of I27 for all frames of the loaded trajectory. The structure corresponding to the red highlight rectangle in the Timeline (left) data panel is shown in the 3D view (right panel). See Figure 9 for a magnified view of the color key.
|
|
- Get detailed information about the trajectory. Details about the residue beneath the 2D cursor are shown in the details box to left and below the box plot. The current residue number, residue name, chain, segment, and frame, as well as the data value for the selected residue, and the current analysis method. These change as you move the cursor.
- Zoom and scroll around the data plot. Right-mouse-click-and-drag to
draw a green-outline box to define the area to zoom into. Right-click will
zoom out. The three slider controls on the right side of the Timeline window
also control zoom: the vertical slider zooms vertically, the horizontal slider
zoom horizontally, the central zoom slider scales the whole data block at once.
The fit all button scales both axes to fit within the standard size
of the Timeline window. The every residue button changes the
vertical scale to display numbering for every vertical element (residues or
selections (covered below, in Section 4.3)).
Figure 9:
Color key for secondary structure plots.
|
|
- Examine color scale key: The color key for secondary structure attributes
is labeled ``sec. structure'', and displays the one-letter structure codes used
by STRIDE software. The one-letter code descriptions are listed in Help
 Structure codes..., and in Figure 9. Most
relevant here, `E' (yellow) corresponds to 'Extended conformation', the main
component of beta sheets; `T'(aqua) corresponds to ``Turn'', another beta sheet
component, and `C' (white) stands for ``Coil'' (random coil, no structure).
Note that secondary structure is a special case for coloring; for all other
cases of calculations, the color scale shows a numerical range.
Structure codes..., and in Figure 9. Most
relevant here, `E' (yellow) corresponds to 'Extended conformation', the main
component of beta sheets; `T'(aqua) corresponds to ``Turn'', another beta sheet
component, and `C' (white) stands for ``Coil'' (random coil, no structure).
Note that secondary structure is a special case for coloring; for all other
cases of calculations, the color scale shows a numerical range.
- Identify the time frames when each of the individual
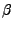 -strands begins to lose secondary structure, and when it has completely lost secondary structure.
-strands begins to lose secondary structure, and when it has completely lost secondary structure.
There are two related ways to help examine how the values of a data set
are distributed: first, by using the Threshold Count tool; second, by
changing the color scaling range of the 2D plot. The Threshold Count tool
adds up, for each frame, the number of residues or selections that fall
within a given range, then plots these numbers for all frames.
The plot is dynamically updated as the Threshold Min and Max values are
changed. Here we review how to do this for the Root Mean Square Fluctuation
data set examined in Figure 4, then use the results from
this to set the color scaling range, in order to identify which structures are involved
in the fluctuation events.
- Display root mean square fluctuation for all frames, all residues in the
trajectory. In the VMD Timeline window, select Calculate
 Calc. RMSF
Calc. RMSF
- Use the thresholding tools. Adjust the Threshold Plot by changing the
Threshold Min and Threshold Max bounds sliders. (Type in values here with
select Threshold
 Set bounds... Set them to 4.48 and 6.67
and note the repeating pattern of peaks. This thresholding can change much
more quickly than we can adjust the 2D plot (especially for a large plot) and
can track changes hard to total up by eye. The Threshold plot updates as the
range is changed, to allow quick exploration of the plots. Now apply the
threshold bounds values we set to the coloring of the whole 2D plot: select
Appearance
Set bounds... Set them to 4.48 and 6.67
and note the repeating pattern of peaks. This thresholding can change much
more quickly than we can adjust the 2D plot (especially for a large plot) and
can track changes hard to total up by eye. The Threshold plot updates as the
range is changed, to allow quick exploration of the plots. Now apply the
threshold bounds values we set to the coloring of the whole 2D plot: select
Appearance Set scaling... and enter 4.48 and 6.67 as bottom and top values. The result should resemble Figure
4c. Scrub the highlight cursor over the transitions in the
resultant plot to see what structures are involved in the fluctuation
increases.
Set scaling... and enter 4.48 and 6.67 as bottom and top values. The result should resemble Figure
4c. Scrub the highlight cursor over the transitions in the
resultant plot to see what structures are involved in the fluctuation
increases.
Perform other changes to the
Timeline plot, to see features that may be helpful when working with different
data sets or different structures:
Figure 10:
Exploring hydrogen bonds during titin I91 extension. The top panel
shows hydrogen bond for all residues of I91 for all frames of the loaded
trajectory. In the middle and bottom rows, structure corresponding to the red
highlight rectangle in the zoomed-in Timeline 2D data plot (left panel) is shown in the 3D view
(right panel). Hydrogen bonds are plotted as white when formed, and black when
broken. The middle row shows highlighted a single, unbroken backbone hydrogen bond. The
bottom row shows highlighted the same backbone hydrogen bond, now broken, at a later time frame.
|
|
Now we will look at the hydrogen bond breaking pattern during the I91
extension trajectory. Since we are looking at the protein architecture, we
will examine the bonds between the partner 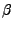 -strands that make up the two
-strands that make up the two
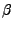 -sheets which form the I91 domain
-sheets which form the I91 domain 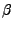 -sandwich.
-sandwich.
- Change the Analysis Selection from all to name N HN O C,
to select only the atoms involved in inter-
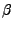 -stand hydrogen bonds.
-stand hydrogen bonds.
- Calculate the hydrogen bonds that can be formed with the current
selection, in Calculate
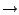 Calc. H-bonds.... Enter a Bond distance cutoff of 3.5 and an Angle cutoff (deg) of 45, then click the Calculate button . The result should resemble Figure 10.
Calc. H-bonds.... Enter a Bond distance cutoff of 3.5 and an Angle cutoff (deg) of 45, then click the Calculate button . The result should resemble Figure 10.
- The value of data in the 2D plot is either 0 or 1. Set the Threshold
Plot range from 0 to 1 to see the number of formed hydrogen bonds in each
frame. (Although the auto-ranging is already set from 0 to 1, you must still
adjust the threshold minimum control once to display the Threshold Plot).
- Zoom in the plot to examine individual hydrogen bonds. Click on the atoms in the 3D view to display the names and numbers of the residues involved.
- Use the hydrogen bond plot, and the fit all button, to look for
overall patterns of hydrogen bonds breaking. Look for H-bond breaking -- the
end of a white horizontal line -- clustered around
the same time, and check the associated structures. While using a copy of a
secondary structure plot such as Figure 8 for reference, note
which
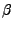 -strands the bond breakings are associated with, and how they relate to the
beginning/ending times of secondary structure loss (unraveling) of individual
-strands the bond breakings are associated with, and how they relate to the
beginning/ending times of secondary structure loss (unraveling) of individual
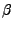 -strands.
-strands.
Figure 11:
Clustered bond breaking during I91 extension. a) Force-extension curve of I91 extension, early force peak is marked with an arrow b) Before clustered bond breaking c) After clustered bond breaking.
In b) and c) , structure corresponding to the red
highlight rectangle in the zoomed-in Timeline 2D data plot (left panel) is shown in the 3D view
(right panel). Hydrogen bonds are plotted as white when formed, and black when
broken. The selected hydrogen bond is highlighted in green in the 3D plots.
|
|
- Load in a version of the I91 extension trajectory with four times as many frames (four times the temporal detail):
navigate to to the examples directory and load titin-big.dcd
- We will examine a set of inter-
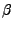 -strand hydrogen bonds that are stable during equilibration of I91. Change directory to the examples directory and type
-strand hydrogen bonds that are stable during equilibration of I91. Change directory to the examples directory and type
source stable-titin-backbone-hbonds.tcl in the Tk console. This will add two vmd atom macros: backboneHSel and backboneOSel.
- Change the analysis selection to backboneHSel or backboneOSel.
- Select Calculate
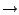 Calc. H-bonds... and click the Calculate button.
Calc. H-bonds... and click the Calculate button.
- Scrub through the trajectory with the highlight cursor, using the Threshold count tool to note how the hydrogen bond count has the largest drop between frames 11 and 18. Note where these breaking H-bonds are in the 2D view, as in the previous example.
- Add both CPK and Hbond graphic representations as for selection
backboneHSel or backboneOSel. Note the hydrogen bonds ending in the
2D graph between frames 11 and 18, and how they correspond to the hydrogen
breaking in the 3D view, as shown in Figure 11. These
simultaneous hydrogen bond breaking seen in Figure 11c is
what is responsible for the extension force peak seen in
Figure 11a. Under forced extension, I91 architecture
cannot allow further unraveling until these bonds are broken.
These features help with saving and loading completed calculations (so you won't
have to spend time calculating them again), and with creating and browsing sets
of pre-computed calculations.



Next: Analysis Scripts
Up: Timeline Tutorial
Previous: Interface and controls
Contents
school@ks.uiuc.edu
![]() -strands of the domain's
-strands of the domain's ![]() -sheets, to get more insight into how
the protein architecture handles applied force.
-sheets, to get more insight into how
the protein architecture handles applied force.
![]() -sandwich then unravels as the force extension
takes place, but there is more to the story; we can make a 2D data plot to
examine the fate of each
-sandwich then unravels as the force extension
takes place, but there is more to the story; we can make a 2D data plot to
examine the fate of each ![]() -strand.
-strand.
![\includegraphics[width=3.0in]{FIGS/timeline-titin-ss}](img13.gif)
![\includegraphics[width=1.5in]{FIGS/timeline-titin-ss-gl}](img14.gif)
![\includegraphics[width=4.7in]{FIGS/hbond-break}](img16.gif)
![]() -strands that make up the two
-strands that make up the two
![]() -sheets which form the I91 domain
-sheets which form the I91 domain ![]() -sandwich.
-sandwich.
![\includegraphics[width=4.7in]{FIGS/hbond-details}](img17.gif)