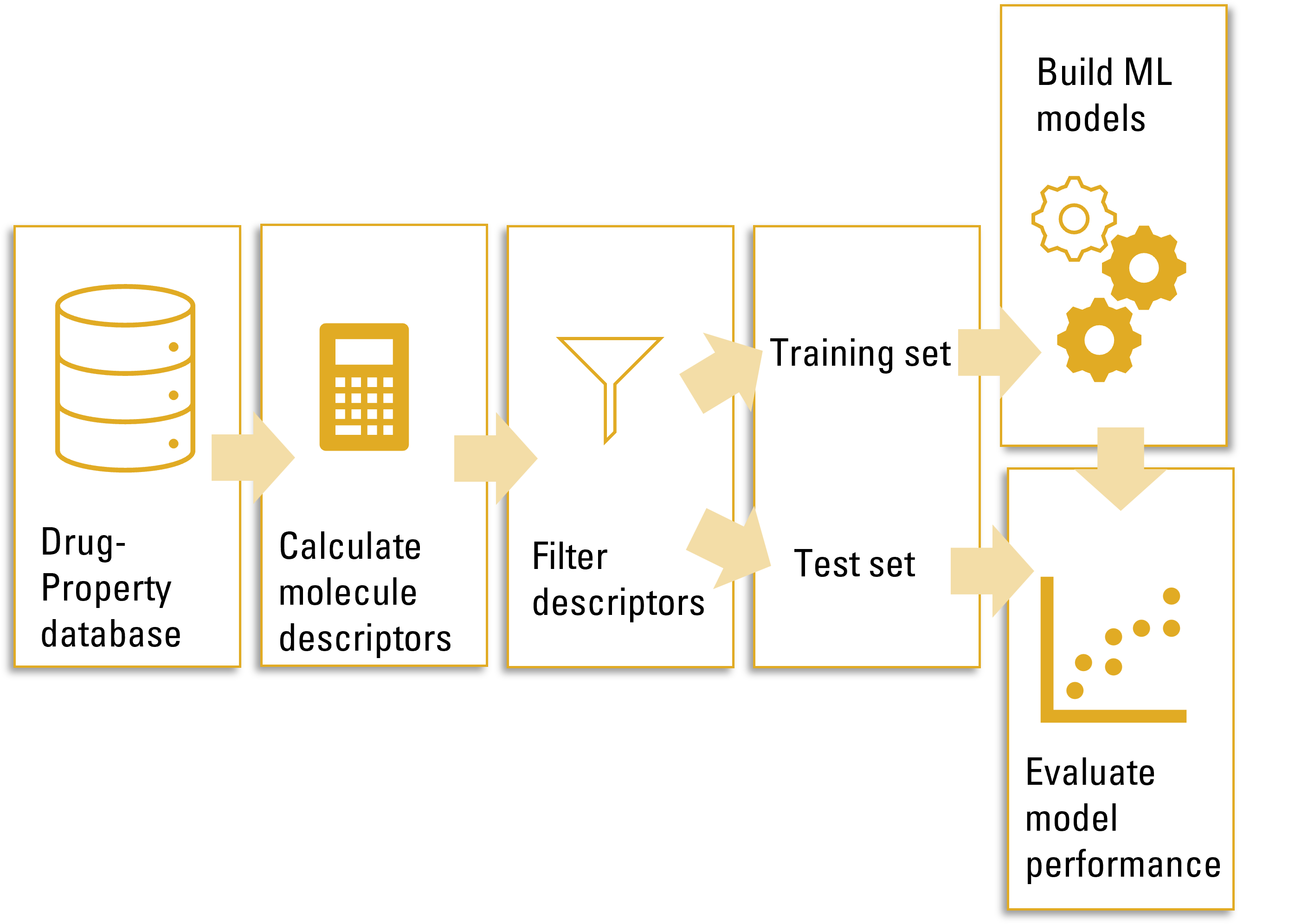
QSPRpred is open-source software libary for building Quantitative Structure Property/Activity Relationship (QSPR/QSAR) models developed by Gerard van Westen's Computational Drug Discovery group. It provides a unified interface for building QSPR models based on different types of descriptors and machine learning algorithms. We developed this package to support our research, recognizing the necessity to reduce repetition in our model building workflow and improve the reproducibility and reusability of our models. In making this package available here, we hope that it may be of use to other researchers as well. QSPRpred is still in active development, and we welcome contributions and feedback from the community.
QSPRpred is designed to be modular and extensible, so that new functionality can be easily added. A command line interface is available for basic use cases to quickly, explore varying scenarios. For more advanced use cases, the Python API offers extra flexibility and control, allowing more complex workflows and additional features.
Internally, QSPRpred relies heavily on the RDKit and scikit-learn libraries. Furthermore, for scikit-learn model saving and loading, QSPRpred uses ml2json for safer and interpretable model serialization. QSPRpred is also interoperable with Papyrus, a large scale curated dataset aimed at bioactivity predictions, for data collection. Models developed with QSPRpred are compatible with the group's de novo drug design package DrugEx.
QSPRpred can be installed with pip like so (with python >= 3.10):
pip install qsprpredNote that this will install the basic dependencies, but not the optional dependencies. If you want to use the optional dependencies, you can install the package with an option:
pip install qsprpred[<option>]The following options are available:
- extra : include extra dependencies for PCM models and extra descriptor sets from packages other than RDKit
- deep : include deep learning models (torch and chemprop)
- chemprop: include the ChemProp integration (only ChemProp versions < 2.0.0 supported at the moment)
- full : include all optional dependecies (requires cupy,
pip install cupy-cudaX, replace X with your cuda version)
If you plan to optionally use QSPRpred to calculate protein descriptors for PCM, make
sure to also install Clustal Omega. You can get it via conda (for Linux and MacOS
only):
conda install -c bioconda clustaloor install MAFFT instead:
conda install -c biocore mafftThis is needed to provide multiple sequence alignments for the PCM descriptors. If
Windows is your platform of choice, these tools will need to be installed manually or a
custom implementation of the MSAProvider class will have to be made.
After installation, you will have access to various command line features and you can use the Python API directly ( see Documentation). For a quick start, you can also check out the Jupyter notebook tutorials, which document the use of the Python API to build different types of models. The tutorials as well as the documentation are still work in progress, and we will be happy for any contributions where it is still lacking.
Contributions and issue reports are more than welcome. Pull requests can be made
directly to the main branch and we will transfer them
to contrib when scheduled for the
next release.