



Next: General parameters and input/output
Up: NAMD 2.7 User's Guide
Previous: External Program Forces
Contents
Index
Collective Variable-based Calculations1
The collective variables module replaces and greatly extends the
functionality of the older freeEnergy module for free energy
of conformational change calculations. The older module is still
available in the code, but has been removed from the documentation to
discourage future use.
In today's molecular dynamics simulations, it is often useful to
reduce the great number of degrees of freedom of a into a few
parameters which can be either analyzed individually, or manipulated
in order to alter the dynamics in a controlled manner. These have
been called `order parameters', `collective variables', `(surrogate)
reaction coordinates', and many other terms. In this section, the
term `collective variable' (shortened to colvar) is used, and
it indicates any differentiable function of atomic Cartesian
coordinates,
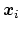 , with
, with 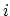 between
between 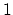 and
and 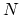 , the total
number of atoms:
, the total
number of atoms:
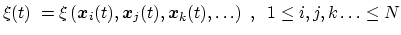 |
(11) |
The colvars module in NAMD offers several features:
- define an arbitrary number of colvars, and perform a
multidimensional analysis or biased simulation by accessing any
subset of colvars independently from the rest (see
9.1);
- combine different functions of Cartesian coordinates (herein
termed colvar components) into a colvar defined as a
polynomial of several such components, thereby implementing new
functional forms at runtime; periodic, multidimensional and
symmetric components are handled transparently (see 9.2.1);
- calculate potentials of mean force (PMFs) for any set of
colvars, using different sampling methods: currently implemented are
the Adaptive Biasing Force (ABF) method (see
9.3.1), metadynamics (see
9.3.2), Steered Molecular Dynamics (SMD) and
Umbrella Sampling (US) via a flexible harmonic restraint bias (see
9.3.3);
- calculate statistical properties of the colvars, such as their
running averages and standard deviations, time correlation
functions, and multidimensional histograms, without the need to save
very large trajectory files.
Subsections




Next: General parameters and input/output
Up: NAMD 2.7 User's Guide
Previous: External Program Forces
Contents
Index
namd@ks.uiuc.edu
![]() , with
, with ![]() between
between ![]() and
and ![]() , the total
number of atoms:
, the total
number of atoms: