Extending a Model¶
In the previous calculations, we used only Cα atoms for calculating normal modes. Publication-quality graphics that use VMD’s cartoon or ribbon representations require a least the full protein backbone. In this part, we will use NMWiz’s Extend model to capabilities to extend NMA results to all backbone atoms of the protein.
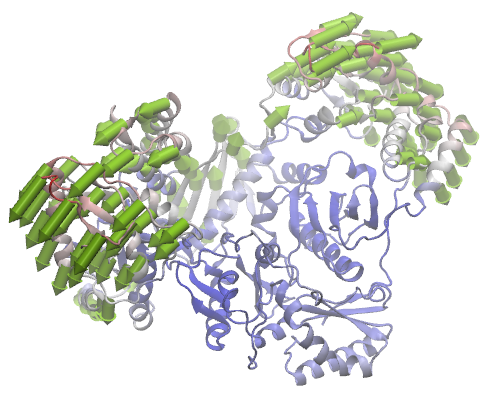
ANM mode 1 for HIV Reverse Transcriptase
ANM Calculation¶
Let’s fetch an X-ray structure of the protein HIV reverse transcriptase (RT) and load into VMD:
$ prody fetch 1dlo
$ vmd 1dlo.pdb
In the ProDy Interface, we select ANM Calculation, check backbone option, and click Submit Job. Model will be calculated for 971 selected Cα atoms, but the normal modes will be extended to all backbone atoms.
Visualization¶
When the results are loaded, you will see four arrows per residue (or node).
Change the Selection string to read name CA and click
Redraw. This will draw only one arrow per residue.
RT is a large structure and updating the display with every little change you make might be time consuming. You can uncheck auto update graphics option in Mode Graphics Options panel to speed up rendering.
To get the view displayed in the figure, you will need to hide arrows that
are shorter than a given length using Draw if longer than option
in the Model->Options menu. Draw an arrow for every forth
residue using the selection
name CA and resid % 4 == 0. The protein representation is NewCartoon.
Animation¶
You can generate a trajectory along the selected mode by clicking Make in Animation row. For large proteins, keeping the Graphics resolution low (10) will make the animation run smoother.
